Improve the quality of your SPR data with these optimization strategies
If you aren’t familiar with Surface Plasmon Resonance (SPR),
check out our article on What is SPR? before continuing.
What is non-specific binding?
During an SPR experiment, there are many experimental parameters that must be controlled in order to optimize the performance of the system and to generate high-quality binding data. Of these parameters, minimizing non-specific binding (NSB) is one of the most imperative as it can directly affect the accuracy of your kinetic data. SPR experiments typically consist of a ligand – the biomolecule immobilized on the sensor surface, and the analyte – the solubilized biomolecule that binds the ligand. The analyte is flowed over the immobilized ligand, and as they interact there is a change in refractive index at the sensor surface. This change in refractive index is measured as response units (RU), which is then used to calculate the affinity and kinetics of the interaction. In some cases there can be non-specific interactions between the analyte and non-target molecules on the sensor surface, which can inflate the measured RU and lead to erroneous calculated kinetics. This is known as non-specific binding.
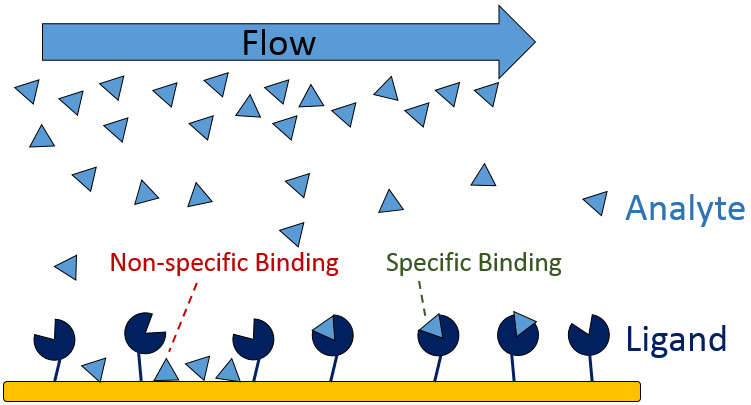
Figure 1: Non-specific and specific binding in an SPR experiment.
How does non-specific binding occur?
NSB is caused by molecular forces between the analyte and sensor surface that can take the form of hydrophobic interactions, hydrogen bonding, Van der Waals interactions, and other nonspecific interactions. The occurrence of NSB in a particular experiment can be the result of several factors, such as the biomolecular coating on the sensor surface, the chemistry used to immobilize the ligand, or a conformational change of the ligand during immobilization. Fortunately, there are many strategies that can be applied to reduce any observed NSB, but for maximum effectiveness it is imperative that you determine the level of NSB before you start the experiment.
Strategies to reduce non-specific binding
One simple preliminary test that can be done is running the analyte over the bare sensor surface without any immobilized ligand. If you observe a significant level of NSB, there are a variety of steps that you can take to limit these interactions. Common strategies include:
- Adjusting the pH of your buffer.
- Using protein blocking additives.
- Adding non-ionic surfactants.
- Increasing salt concentration.
In order to select the best possible method, it is important to know the characteristics of your analyte and ligand. Knowing the isoelectric point, charge, size, and composition (hydrophilic, hydrophobic) of the molecules can help you determine the most effective condition(s) for reducing NSB in your experiment.
1. Adjust the pH of your buffer
The pH of the running buffer and the buffer in which your analyte is solubilized can have a significant impact on NSB because it dictates the overall charge (positive or negative) of your biomolecule. For example, if you are using a pH at which your analyte is positively charged, the analyte will non-specifically interact with a negatively charged sensor surface. In such a case, you have the option of adjusting your buffer to a pH within the isoelectric point range (predicted neutral overall charge) of your protein, or neutralizing the surface of the sensor.
2. Use Buffer Additives – Protein Blocker
If you are using a protein as your analyte, a good first step to preventing non-specific binding is adding bovine serum albumin (BSA), a commonly used protein blocking additive, to your buffer and sample solution. As a globular protein composed of domains with varying charge densities, BSA can surround your analyte to shield it from non-specific protein-protein interactions, interactions with charged surfaces, glass and plastic surfaces. Thus, additives such as BSA can be used to minimize NSB as well as prevent analyte loss to tubing in your system. BSA is usually used at a concentration of 1%, but can vary from experiment to experiment.
3. Add Surfactants – Tween 20
If non-specific binding is occurring due to hydrophobic interactions in your system, it may be beneficial to use non-ionic surfactants such as Tween 20. Low concentrations of this mild detergent can disrupt hydrophobic interactions between the analyte and sensor surface. Similar to BSA, Tween 20 is also commonly added to prevent the analyte from binding to tubing and container walls.
4. Increase salt concentration (NaCl)
In systems where NSB occurs primarily due to charge-based interactions, using higher salt concentrations in your buffer can reduce these interactions by producing a shielding effect on the analyte. Salts such as NaCl can be used at varying concentrations to prevent charged proteins from interacting with other charged surfaces, including the sensor surface.
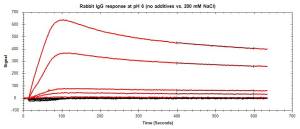
Figure 2: Reduction in NSB of a charged analyte (rabbit IgG) with the addition of 200 mM NaCl. Red curves represent non-specific binding from various analyte concentrations. Black curves represent the corresponding response of analyte with the addition of salt to the running buffer.
For a detailed experimental analysis of these methods, download our application note.
Optimize performance by planning ahead
Preventing non-specific binding (if it exists) is key to performing successful SPR experiments. The more information you have on your ligand and analyte, the easier it will be to optimize experimental conditions and minimize NSB. While there are strategies that can be applied in any given case, it is important to note that extreme conditions may deactivate or denature your biomolecules. As such, always have in mind the potential effects on your biomolecules when deciding on an intervention to prevent NSB. If you find that you are unable to eliminate all NSB from your system, but the quantity of specific binding is much greater than the NSB, subtracting the NSB signal from the specific binding event can help alleviate the impact of NSB on your data.
By correcting for non-specific binding in your SPR experiments, you will gain confidence in the quality of your data and be able to accurately analyze the biomolecular interactions that you are interested in. This will enable you to better understand your biological system of interest. At Nicoya, it is our goal to improve human life by helping scientists succeed, and helping to optimize experiments is just one of the ways we accomplish this mission. For more optimization tips, check out our blog post on Improving SPR Data Quality. If you have any questions about reducing NSB or would like to learn about how SPR can be applied to your research, we’d love to help!

